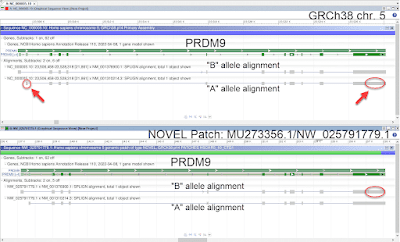
Since the release of GRCh38, the GRC has received a number of user reports alerting us to a potential false duplication involving chr 21p and 21q. Users noted that reads were aligning to both regions in GRCh38, but not GRCh37/hg19, resulting in a decreased mapping score and difficulties in variant calling throughout. Additionally, user analyses involving Multiplex Ligation-dependent Probe Amplification (MLPA), a technique for gene copy number detection, and exome studies indicated potential false duplications. The implicated regions contained several genes, including CBS (Gene ID: 875),
U2AF1 (Gene ID: 7307) and KCNE1B (Gene ID: 3753). The GRC has investigated the matter and concurs that the GRCh38 assembly contains sequence on the short arm of chr 21 that should be excluded from analyses. Read on to learn more about this issue, as well as some recently detected non-human contamination in GRCh38, and ways you can find and avoid these sequences in your analyses.
The short arm of human chromosome 21, like that of the four other human acrocentric chromosomes, is where genes associated with rDNA synthesis are localized, and is characterized by highly repetitive heterochromatic sequence. The repetitive nature of these sequences, coupled with limitations in sequencing technology, have until recently made the representation of these regions in genome assemblies very difficult.
As a consequence, the GRCh37 representation of the chromosome 21 p-arm contained only 11 clone sequences. Seven were clones from the HSA21-specific BAC library CHORI-507 that had previously been experimentally localized to 21p (PMID: 17895424). In an effort to add additional sequence to this repetitive region, 23 additional components were added to 21p for GRCh38, including 18 additional CHORI-507 clones, 4 RPCI-11 clones, and 1 ABC9 fosmid. Admixture mapping localized some of these clones to this region.
In response to the user reports, the GRC re-reviewed the sequences added to 21p in GRCh38. Haploid CHM13hTERT Illumina reads generated by The McDonnell Genome Institute were aligned to GRCh38 by NCBI, and evaluated for read mapping and coverage. This analysis supported the user reports, suggesting that 5 of the newly added CHORI-507 clones (FP565260.4, CU639417.17, FP236240.8, FP475955.4 and CU633980.13) were actually redundant with sequences on chr 21q, and thus represented false duplications in GRCh38.
The GRC has now removed these sequences from the files that it uses to generate the reference assembly. However, we cannot remove them from the GRCh38 assembly without triggering the next major release of the human assembly. In order to help users recognize these regions and avoid them in their analyses, we have produced a masking file to be used as a companion to GRCh38. This BED file is available from the GenBank FTP site: https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/405/GCF_000001405.39_GRCh38.p13/GRCh38_major_release_seqs_for_alignment_pipelines/GCA_000001405.15_GRCh38_GRC_exclusions.bed. This file provides the assembly coordinates of the 5 clones incorrectly localized to chr 21p. The Genome in a Bottle Consortium recently posted a preprint demonstrating that using this masking file greatly improves variant calling accuracy in the affected genes (https://doi.org/10.1101/2021.06.07.444885).
In addition to these sequences, the file also includes 2 other assembly scaffolds that were found, after the release of GRCh38, to be contaminated with non-human sequence. These include a chr Un scaffold (KI270752.1/NT_187507.1), whose sole component (AF065393.1) is now known to represent sequence from Chinese hamster (PMID:30486838), likely derived from the human-hamster CHO cell line that was the clone source, and an alternate loci scaffold (KI270825.1/NT_187580.1) whose non-anchor component (AC225822.3) was shown to be chimeric. In AC225822.3, the first 25,375 bases are human sequence matching GRCh38 chr 10 reference component and alternate scaffold anchor sequence AL391421.27, while the rest match Acidithiobacillus thiooxidans sequences from multiple WGS projects (PMID:32398145). Although all sequences in the reference assembly are screened for foreign contamination, these two were not detected at the time of release (2014). Prompted by these findings, the GRC has more recently re-screened the assembly with updated contamination databases and has not detected additional issues. As these two scaffolds are not human sequence, very few reads are likely to map well to them, but users may still want to make note of them in their analyses.
In total, the contamination represents ~800 Kb, or 0.02% of the total sequence length. GRCh38 remains an extremely high quality reference assembly. Nonetheless, the GRC remains committed to addressing assembly errors and making sure it serves as the most reliable analysis substrate possible. Check out our website to see other genomic regions under review. We welcome your feedback and reports of newly discovered issues! In the future, we plan to update the masking file with any new regions as identified and reviewed by the GRC.
Fig. A
 |
Fig. A: Aligned CHM13hTERT Illumina reads viewed in Integrative Genomics Viewer (IGV). The
panes labeled 'Original' were reads aligned prior to redundant sequence
masking and the panes labeled 'Fixed' are reads aligned after redundant
sequence masking. The top two panes show reads aligned to the
valid U2AF1 locus in GRCh38 (NC_000021.9:43,091,000-43,110,000 of 21q)
and the bottom two panes show reads aligned to the falsely duplicated
(pseudo region) region of GRCh38 (NC_000021.9:6,480,000-6,500,000 of
21p).
In IGV,
sequence reads that align to 2 places in the Reference (whether it is
correct or not), yield poor/ambiguous alignments, indicated by clear,
unshaded reads. This is shown by both the 'Original hg38 U2AF1' and the
'Original hg38 pseudo region' pane. Following the masking of the
known, duplicated region introduced in GRCh38, the aligned reads in the
'Fixed hg38 U2AF1 gene' pane are shaded grey, meaning they have good
mapping scores to that region. And there are no reads mapping to the
'Fixed hg38 pseudo region' because the duplicated sequence is masked in
the Fixed hg38 file. |
Fig. B
 |
| Fig. B: Aligned reads to 21q region that has false duplication on 21p in
GRCh38 before masking. Note the BAC clone boundary where alignment of
falsely duplicated region in 21p starts. This duplication involves the
CBS gene (Gene ID: 875). |
Fig. C
 |
| Fig. C: Aligned reads to 21p region falsely duplicated (in Fig. B). You
can see ambiguous read alignment and the falsely duplicated CBS gene
annotated in the gene track. |
Fig. D
 |
| Fig. D: Alignment of BAC FP236240.8 (redundant BAC added to 21p for
GRCh38) to the corresponding valid region on 21q. Note the redundant BAC
alignment to the region (bottom pane) and the valid read alignment
depth (shown in middle pane). Since the region was falsely duplicated,
read alignment in the region of redundant BAC alignment is poor. |